As its name suggests, CAD tools help us design stuff, normally in 3D. To explain why these kinds of tools are so useful, imagine the work of a person who designs airplanes, skyscrapers, or computers. These things are quite complex, and so drawing everything by hand, isn’t longer feasible.
Now, what if I told you that biology can also be designed? This is the art of synthetic biology, also called biological engineering.
Let’s take a look into how CAD is used in this field…
Engineering Biology
Just in case that you aren’t very familiar with synthetic biology, I definitely recommend that you check out this guide to it.
The reason we would want a software that designs biological stuff, is because in the XXI century, we want to standardize life. We want to better predict what cells will do, and overall we want to think of them more as computers rather than burritos.
If you agree with me, computers are logical. They are literally based off of things called logic gates. This means that they follow simple logic in which 0 will always be different from 1.
Just a few decades ago, we used to think that this didn’t happen with biology, just because of the innumerable amount of interactions that occur every single second. However, the rise of biological engineering has brought us to think the opposite.
Genetic circuits
Research on biological computing is still going on, and so talking about DNA logic gates is actually kind of impressive. Further in this article I’ll go over the tool that I used to design these logic gates.
For now, it’s important to understand how these actually work in the world of biology.
In this context, our inputs could be substances such as DNA, in the form of genes, and our outputs would be proteins. This means that if a certain gene is active, it could lead to the production of a certain protein, depending on the logic gate that we’re talking about.
AND Gate
If the two inputs are true, the output will also be true.
Inputs: two small molecules called inducers
Inducers: controls the binding of the regulatory protein to the specific binding site on DNA
Operator part: DNA containing a protein-binding site
As of this specific image the specific components are:
Pale blue arrow: promoter
Orange rectangle: downstream operator part
Blue ellipse: a ribosome-binding site
Black arrows: environmental inputs
Gray arrow: coding region (output)
NOT Gate
The output is only produced in the absence of the repressor. Otherwise, the presence of the repressor obstructs the bound of RNAp and promoter.
Input: a repressor.
Repressor: DNA- or RNA-binding protein that inhibits the expression of one or more genes
Another explanation of this gate would be:
A NOT gate is a promoter that is turned on because of an environmental condition. This promoter then turns on an inhibitor, which inhibits another promoter, which would have activated a gene otherwise. In other words, when the input is 1 (first promoter is activated), then there will be no output (the last gene won’t be active)
A NOR gate would be a very similar one, only that if any of the two first promoters are activated, then the output will be 0.
Here are some examples of other logic gates:
Increasing complexity
The way I like to see it, these gates can then arranged to form more complex systems, which are circuits.
Just to learn a bit more about this field, I decided that my first project would consist of creating a genetic toggle switch. What is this?
Well, it’s not that big of a deal in electronics now. In fact, it’s everywhere on our smartphones’ screens. Once a toggle switch is ON, it remains ON until you press the switch again.
Now, the logic behind is a little more complex than it looks like. It is composed of two repressors and two constitutive promoters. Each promoter is inhibited by the repressor that is transcribed by the opposing promoter.
The strength of each can be enhanced by adding aTc or IPTG to the cellular environment.
The levels of LacI and TetR (repressors) can be observed using two fluorescent reporters.
Synthetic Biology Open Language
CAD software is used to provide multimedia representations of circuits through images, text and programming language applied to biological circuits. This makes the design step improve more rapidly by giving more organization to gene interactions and mathematical models.
A pain in the neck?
On the subtitle you can read though, that these tools can be a lifesaver as much as a mess these days. Well, yes. They can help a lot… when you’ve downloaded them and when they’re at least a little bit updated.
From a surface level perspective, I noticed that a lot of the applications listed on the SBOL page are outdated. It looks as if the websites had only been created for a high school project, and then their creators forgot about them.
Cello, the synbio CAD, not the musical instrument, is supposed to be one of the best tools out there to design biological circuits. Its purpose is to allow users to program biological computers, just as they’d program conventional computers: using code.
Cello creates a genetic circuit design from the Verilog code. It does this via a set of algorithms that parse the Verilog text, create the circuit diagram, assign gates, balance constraints to build the DNA, and simulate performance
Well, I don’t have a doubt that this once worked, but when I tried to use it, the program didn’t allow me to upload my DNA files, nor did it let me define what I wanted the inputs and outputs to be.
Then, there is Tinkerbell. I mean… Thinkercell. Some friends of mine had already used this program, and they did well. I wanted to do that too, but apparently it doesn’t support Mac these days.
Finally, I found out that there’s a tool called SBOL Designer. I’m pretty sure that having trouble with this one was my fault, since I got confused when downloading Java. Still, I think developers could simplify this a lot more.
After some time using SBOL, I noticed that when I wanted to actually download a genetic sequence from a database, it wouldn’t work. If the program can’t do that, it basically means that it’s not much more useful than drawing the designs by hand.
A lifesaver?
Yes, I ended up using SBOL for my project. So, once I managed to make the program work, figuring out how to use it wasn’t that big of a deal. There are many buttons you can use, but each one of them has a very clear function. You just drag and drop blocks, it’s really that simple.
On the positive side, SBOL allows you (supposedly) to not only have a clear view of your synbio design, but also have different abstraction levels, work with a variety of biological parts, and assign actual DNA sequences to each one of them that you can import from databases, including ones from iGEM’s repository.
Toggle switch
On the project itself (finally), I can say that it was kind of complicated to understand the different levels of abstraction at the beginning (especially being new to the field). However, after trying it out at different times with different files, I managed to build something.
The toggle switch that I created is exactly the same as the one I described above. It is composed of 2 main parts that repress each other.
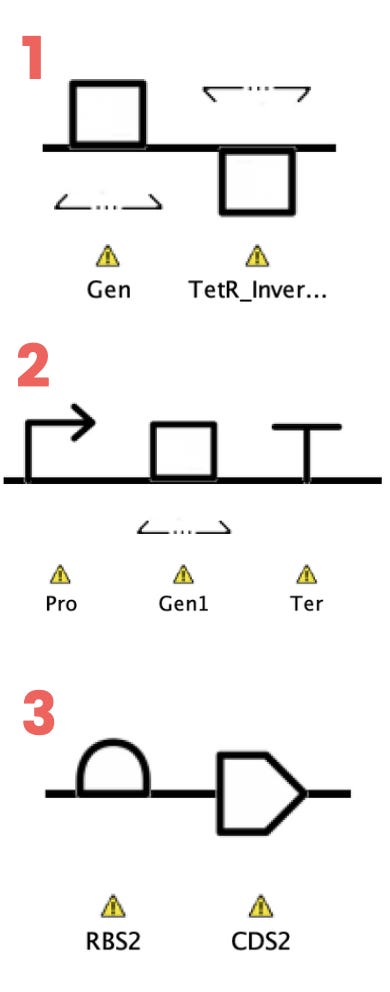
When TetR is active, it represses LacI and vice-versa. Each of them is composed of a promoter, a gene, and a terminator. The coding sequence at the same time, is composed of a Ribosome Binding Site (RBS) and a Coding Sequence (CS).
Explained differently, the levels of abstraction are:
Toggle switch
LacI and TetR repressors
Promoter, gene, terminator
Gene: RBS, coding sequence
The image on the left shows how that looks like on SBOL Designer. Note that the yellowish triangles are there because of the error with assigning a DNA sequence to the parts.
In the end…
This small project was challenging at the same time as it was simple. At first, I didn’t even know what kind of circuit I wanted to create, or which program to use, and it was also hard to actually download those programs, but once I did, I was able to see how these CAD systems can greatly simplify the design process in genetic engineering.
The toggle switch is just one example of what can be achieved when we want to emulate circuit logic in biology. By continuing to increase the complexity of our systems, we will be able to create biological computers that treat diseases, make calculations, or store information to solve the world’s biggest problems.
IT’S TIME TO GROW!