Playing Molecular detectives: making an Alzheimer's drug by discovering new metabolic pathways in daffodils
Explained version of pre-print paper by Niraj Mehta, Yifan Meng, Richard Zare, Rina Kamenetsky-Goldstein, and Elizabeth Sattely
Background
Analogous to contemporary technologies like cryopreservation of oocytes or our smartphone’s rest mode, plants and use dormancy as a survival strategy that responds to environmental cues. As their name implies, geophytes like Daffodils are a group of plants that can store their resources underground or under water—in organs such as bulbs, tubers, corms, or rhyzomes—before re-germinating.
These plants tend to produce chemical toxins to defend their food reserves from herbivores, one example being the organosulfur flavor compounds produced by onion and garlic. Such compounds that are not absolutely essential for growth, reproduction and development are termed secondary metabolites. In plants, they are classified by their structure into phenolics (the most abundant type, like Resveratrol), terpenoids (some of which are phytohormones), alkaloids (contain nitrogen), glucosinolates (contain sulfur and nitrogen, derived from glucose).
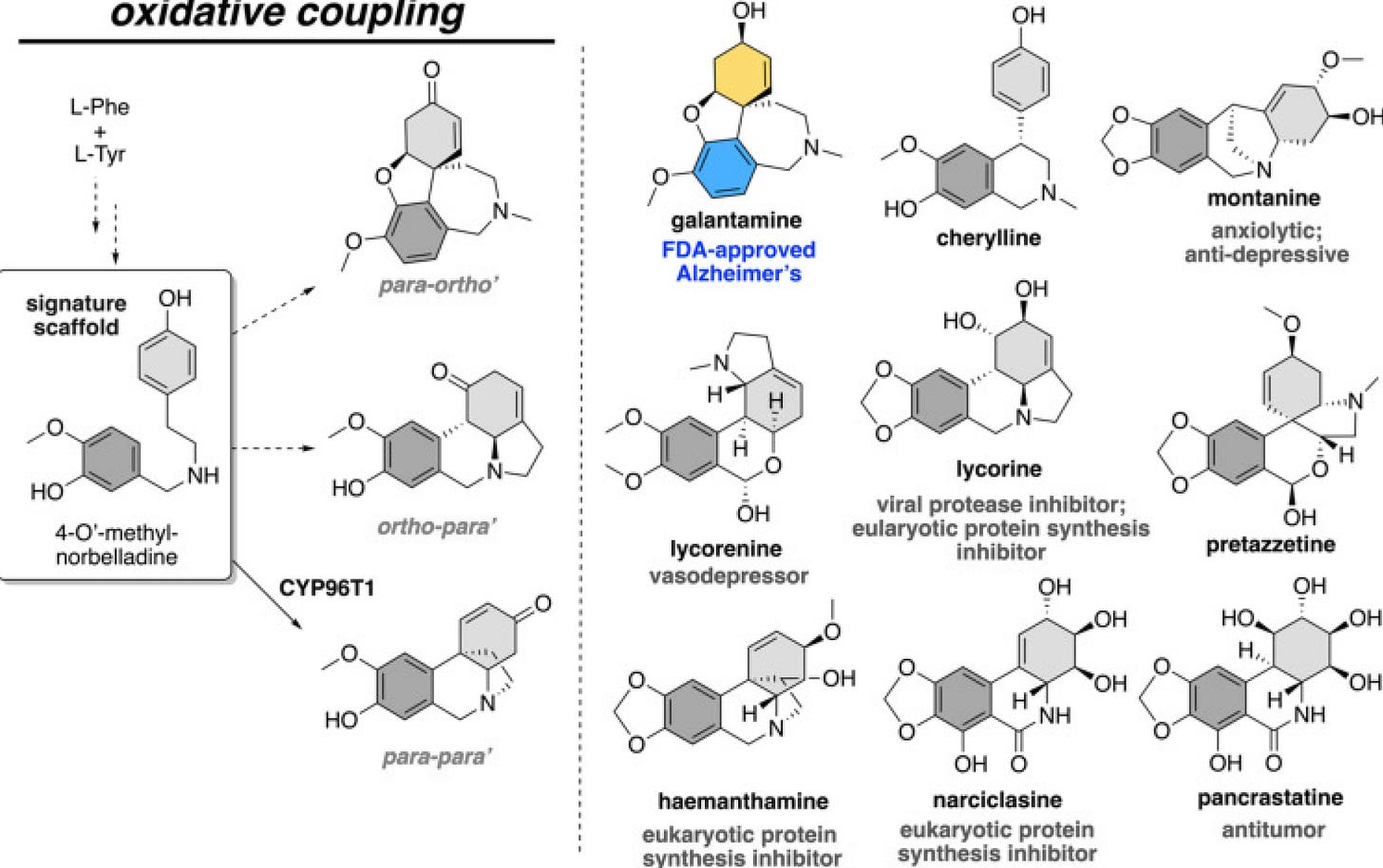
Diversity-oriented biosynthesis tells us how chemical diversity can be generated through iteration of the same precursors. This paper focuses on the set of >150 Amaryllidaceae alkaloids (AmA) that derive from a common precursor, 4-O’-methylnorbelladine (4OMN) through either of 3 pathways (p-o’, o-p’, or p-p’).
Even though the enzyme CYP96T1 has previously been identified for the p-p’ pathway, the enzymes for other pathways were not known until now. In his outstanding PhD thesis work, Niraj set out to uncover the full AmA pathway by:
Identifying where the AmA are actively being synthesized
Finding the key coupling enzymes through expression of the known ones in N. Benthamiana (Benthy for us)
Working backwards from the previous ‘clues’ to uncover the whole pathway, thus enabling the production of that Alzheimer’s drug in simpler organisms through synthetic biology
This is my attempt to #ExplainThisPaper in the easiest and shortest way possible. If you enjoy reading about science like this, subscribe for more:
1. Find molecule production site
In our Molecular Detectives game, Daffodils are a large city where many different molecules live. Each molecule has a very unique history (like an ID) that mentions what enzymes were involved in its production and what other substances are derived from it. If only Niraj had access to the city’s secret database, it would be way easier to find each molecule and its enzyme. Except he does not. 😈
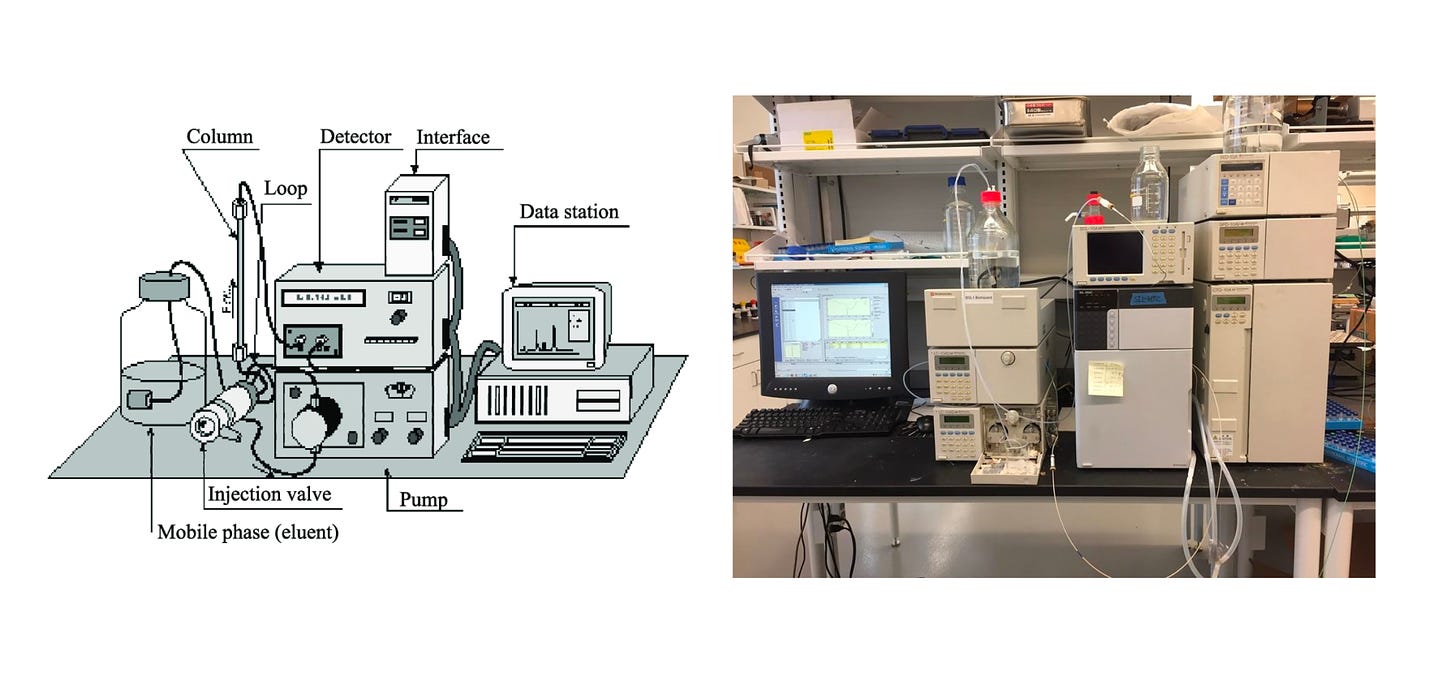
As a molecular detective, Niraj will first have to search for traces of the AmA across different plant tissues using a technology called Liquid chromatography–mass spectrometry (LC-MS), ‘chromatography’ referring to separating things and ‘spectrometry’ to measuring things. LC separates liquids and mass spectrometry measures masses. Since I don’t like LC-MS as a name, I’ll baptize this device as the Molecule Reader from now on.
Before using the Molecule Reader, Niraj needs congregate only the AmA living among thousands of other molecules in Daffodil City. To do this, he may take different slices of the different tissues, grin, vortex and sonicate them to have a homogenous shake, add a solvent that binds to all the other molecules but not AmA, evaporate that solvent, centrifuge the mix to concentrate the AmA, repeat. Until having a concentrated mix.
Roughly, the Molecule Reader works in the following way:
You insert the liquid. Using a pump, the liquid travels through a ‘column’ (i.e. tube), made out of either polar or non-polar materials. If the tube is polar, the more polar AmA will travel slower because they will stop to interact with the polar molecules in the tube. Best video on this step.
The detector can separate the molecules according to different physical properties like UV absorbance, fluorescence, or mass-to-charge ratios. For example, by shining UV light through the compounds and detecting that back with a photodiode, we can match what compounds have a higher or lower absorbance and separate them as such.
Fascinatingly, this photodiode turns photon (light) energy into a proportional amount of electric energy. An Analog-to-Digital Converter is used to turn this electric signal into bits which are stored in binary or text and associated to a time stamp which computer software puts together into a graph or…
The separated molecules can go into the mass spectrometer to measure their abundance in relation to their mass. The substance is ionized (bombarded with electrons) and then deflected using a magnetic field. The lighter the molecule the higher the deflection which is detected in a similar way to the photodiode.
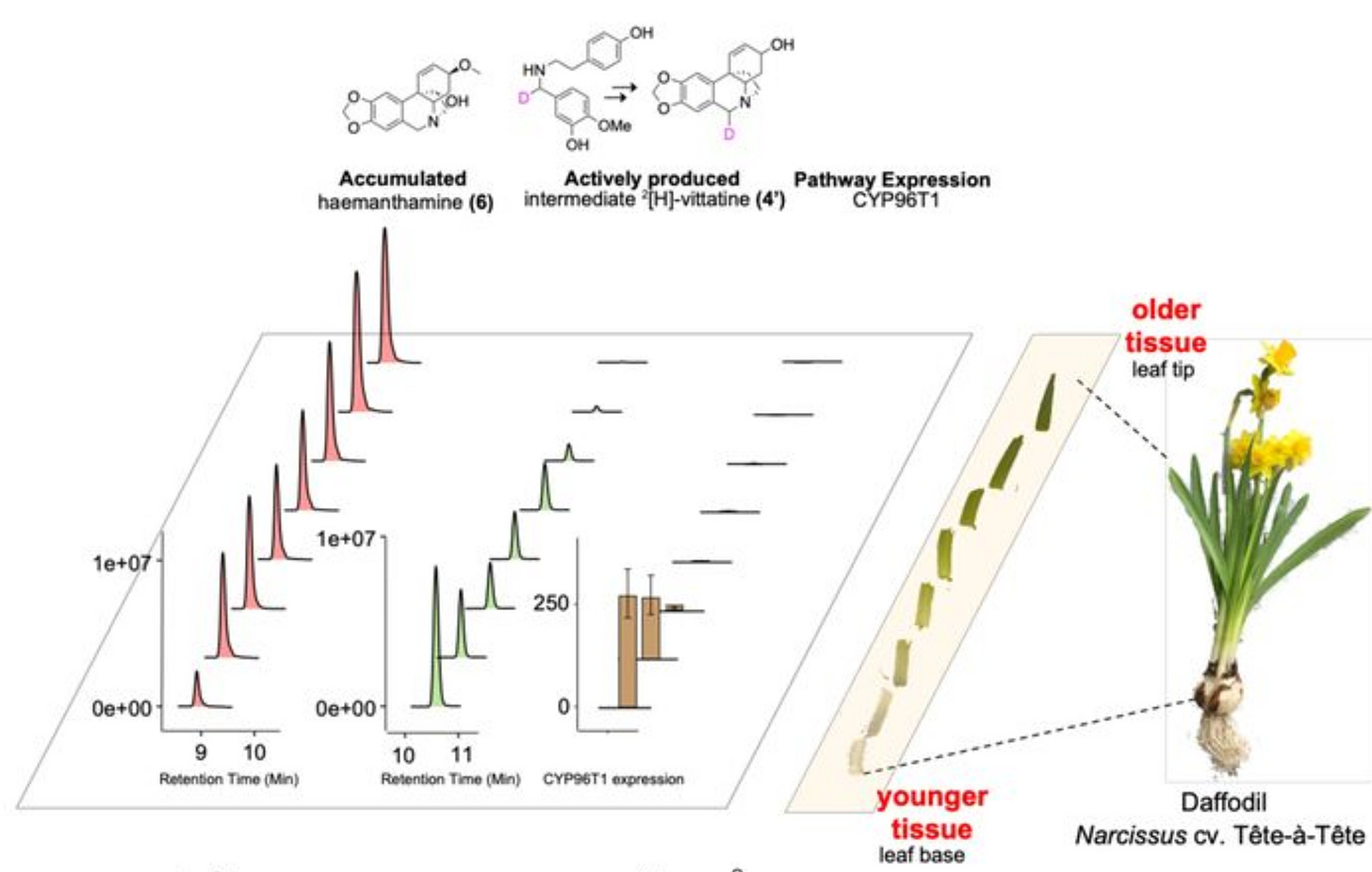
Unsurprisingly, Dr. Mehta only found that AmA are accumulated all around the plant with a slightly greater population in older tissues (tips of the plant). To actually get the precise location for AmA active manufacturing, he set out to follow the trace of the p-p’ pathway intermediate, [2H]-vittatine.
For that, he soaked the plant shake into a solution of isotopically labelled precursor, called [2H]-4OMN with 2H as the label, read the amount of [2H]-vittatine (the precursor) using the Molecule Reader, and found that most of it was in the youngest leaves.
Through this approach, Niraj finally got to the conclusion that AmA are actively synthesized in the youngest leaves and accumulated in the oldest leaves—and the 3 pathways were consistent with the known pattern of the CYP96T1 enzyme expression!
Collectively, this first phase of the game suggests that young leaves are creating these toxins before emerging from the bulb as a means of protection. Could it be that this is the case for other bulb species?
2. Find key enzymes for AmA production
If the DNA is Daffodil City’s secret database, the enzymes are the workers and other proteins are buildings, RNA (the transcriptome) is the construction material. Knowing the active sites of AmA synthesis, Niraj can now read the RNA to identify enzyme candidates to make AmA and he can test those candidates by expressing those enzymes in a simpler organism like Benthy.
This was probably the hardest step in all the study. The molecular detectives thought that the o-p’ enzyme should be related to the already-known CYP96 family but they could not find it after expressing several candidates in Benthy, which could either because of chemical instability or poor protein expression.
The team, however, was sure that this enzyme should be in the same tissue area as the others, so they came up with the seemingly crazy yet marvelous idea of expressing all candidates at once, adding the 4-OMN precursor and using the Molecule Reader to find an intermediate molecule to the AmA.
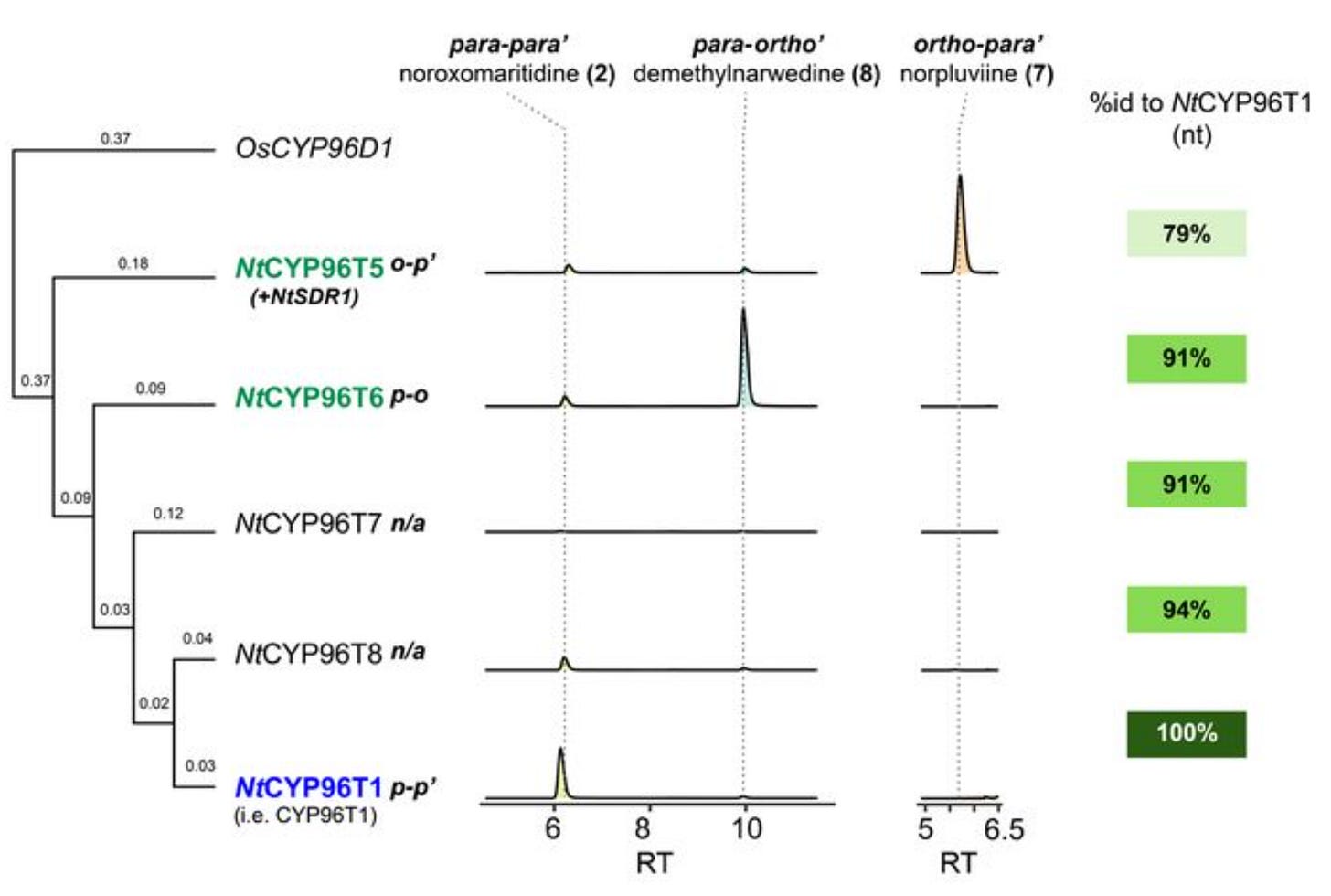
In one one of the tested combinations, they read a highly abundant feature that corresponded to the expected mass of an oxidative product that had undergone a reduction. After further mass spectrometry analysis, and comparing it to a standard, they saw that this was in fact an intermediate in the lycorine o-p’ pathway, norpluviine! Tracking back which of the batch-tested candidates was catalyzing norpluviine, they found that it was not one enzyme but two: NtCYP96T5, a CYP96T1-like P450 and NtSDR1, a short-chain alcohol dehydrogenase/reductase.
To their surprise, p-o’ enzyme (NtCYP96T6) and the o-p’ enzyme (NtCYP96T5) are 91% and 79% similar to the already-known CYP96T1 from the p-p’ pathway, at the nucleotide level—by one single amino acid!! For example, they found that swapping Alanine 308 on CYP96T1 (p-p’ enzyme) to a Leucine like NtCYP96T6’s (p-o’) gave it the ability to catalyze p-o’ coupling—diversity-oriented biosynthesis!
3. Uncover the whole pathway to the Alzheimer’s drug
In biology everything is interconnected to achieve homeostasis (aka balance). To make an Alzheimer’s drug using Daffodils you not only need to know the direct pathway to the drug but also pathways that could inhibit its production. In this case, Niraj and team had to uncover the pathway to haemanthamine (a molecule that can inhibit protein production) before getting to galantamine (the drug).
Fortunately, at this stage they had more clues. They searched for RNA that could encode enzymes involved in haemanthamine production, asked which ones reduce which compounds, expressed them in Benthy with 4OMN and checked if they’re getting the expected molecules using the reader.
In this fashion, they worked backwards from galantamine too, using the information they had about the two new enzymes they’d just discovered in the previous phase. (NtCYP96T6, NtNMT1 and NtAKR1) until hurray: they identified the minimum set of core genes for the biosynthesis of the FDA-approved Alzheimer’s disease therapeutic, galantamine 🥳 !!!
I really wish I could explain the discovery in greater detail but it’s been long… what I can do for y’all though, is tell you that expressing something in plants is a bit of a long process. One can use E. Coli to put different genetic parts together, then transfer that into a popular soil microbe called Agrobacterium which can transfer that DNA into plants like Benthy 🌱 :)
An easy way to verify you did this right is using a gene like RUBY which encodes the enzymes for the synthesis of betalain—yeah, the molecule that makes beetroots red!
An exciting future ahead
Earlier this year I found myself in Monterrey, Mexico, doing metabolomics in avocado to discover anti-microbial compounds that would serve as food conservatives. I was marveled by the fantastic couple that the meta- can be to the geno(mics); how many things are still left to discover and how we can engineer organisms to be manufacturing platforms.
In the future of this field, I’m really excited for what in-vitro platforms and genomes in our pockets could offer. Expanding from our genetic toolkit and learning what specific biochemical buttons to press in plants to elicit the production of the substances we care about, sustainably and at scale.
I have not ceased to philosophize around and advocate for a fair use of our global and local genetic resources. How do we enable more people, without a scientific background, to create with biotech? How do we make sure that those who’ve nurtured our biological heritage for generations, are part of its future too?
Writing is a conversation. I write to understand and attempt to answer questions. I love it when readers keep the conversation going so don’t hesitate to do so, and don’t doubt that you’ll hear back from me soon